| HOME | ABOUT | DOWNLOAD |
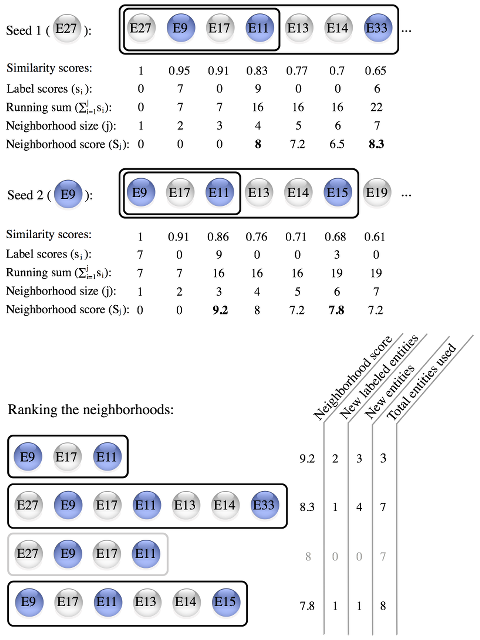
About HOODSClustering algorithms are often used to find groups relevant in a specific context; however, they are not informed about this context. We present a simple algorithm, HOODS, which identifies context-specific neighborhoods of entities from a similarity matrix and a list of entities specifying the context. To produce context-specific groups of similar entities (neighborhoods), we have implemented HOODS, which is a simple algorithm that only needs 2 input files to run: 1) the similarity matrix among entities; and 2) the association scores between entities and the specific context (label). HOODS first sorts the neighbors for each entity in the matrix (seeds). Secondly, for each seed it goes through all the nearest neighbors calculating a neighborhood score:
The parameter α is needed to set how much the user wants to penalize big neighborhoods and it ranges from 0 to 1. In this equation we take into account the label score density in our neighborhoods and the total label score of them. The score is a weighted geometric average from these two components; ideally a neighborhood should have both a high density of labels and a large number of labeled entities. The algorithm sorts all neighborhoods obtained from all seeds by the neighborhood score and filters out neighborhoods that contain the same set of labeled entities as a higher scoring neighborhood. It also removes neighborhoods that contain no entities that are not already included in any of the higher scoring neighborhoods.
Although we group related entities as clustering methods do, HOODS does not intend to be a clustering algorithm. The outcome of clustering is distinct clusters, such that entities in one cluster are more related to each other than to entities in different clusters. Our neighborhoods contain related entities that are grouped in a given context, but some of them may be more closely related to other entities in other neighborhoods. |